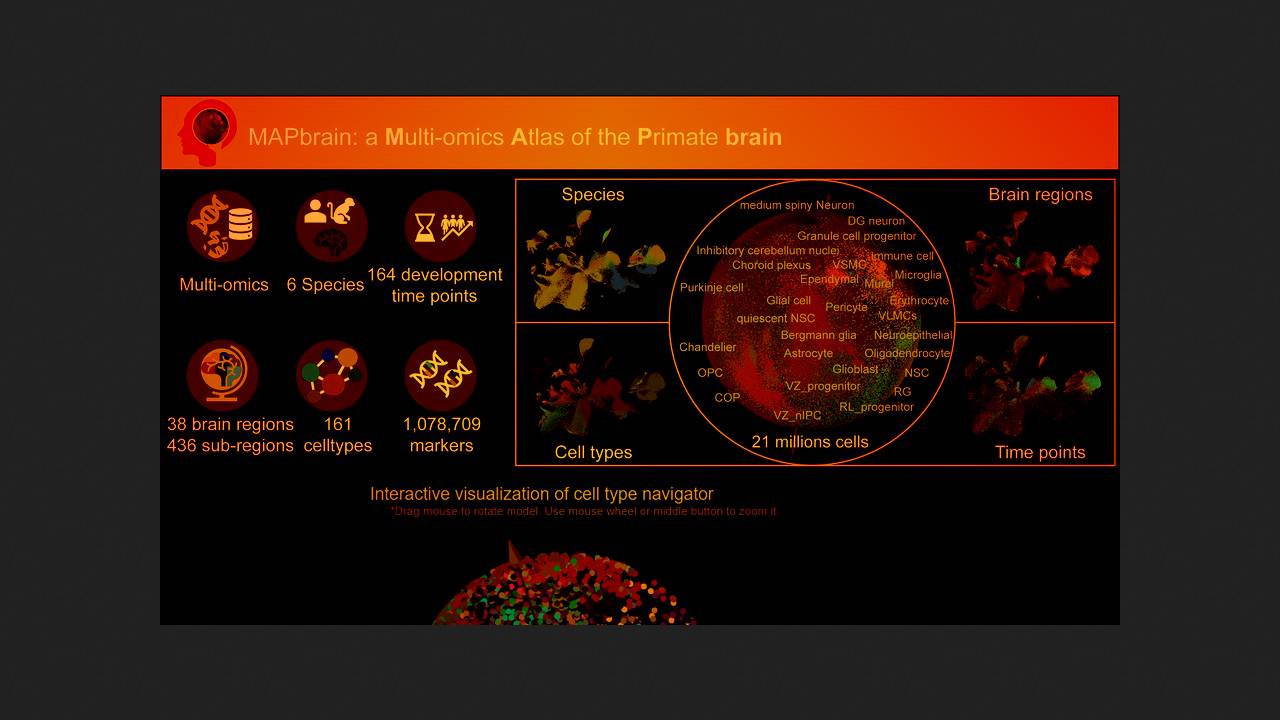
Xiaoqun Wang of the Chinese Academy of Sciences and colleagues presented a multi-ome brain atlas of six primate species, including humans. To create it, the researchers used their own work and the PubMed, Gene Expression Omnibus (GEO), UCSC Cell Browser, NeMO, Descartes, EMBL-EBI, and Allen Brain Map databases to collect and integrate publicly available results from single-cell transcriptomic studies, spatial transcriptomic analyses, and epigenomic analyses of the brain. The resulting gene expression matrices were compared with reference genomes of humans, chimpanzees, gorillas, rhesus macaques, cynomolgus macaques, and common marmosets. The results were published in the journal Nucleic Acids Research.
The analyzed and integrated data were compiled into an atlas called MAPbrain. It contains data on 21 million cells from 38 key regions and 463 subregions of the brain across all periods of embryonic and postnatal development (a total of 164 time points). The atlas can be navigated to study individual cells, the transcriptome, and the epigenome as a whole, to examine molecular markers specific to different cell types and developmental stages, and to compare gene expression profiles across different brain regions and primate species. Users can upload their own data to the online platform and analyze it. The atlas is openly accessible.