US chemists modeled and expressed proteins that bind to porphyrin iron complexes in bacteria and used them to catalytically cyclopropanate double bonds and insert diazo compounds into silicon-carbon bonds. In both cases, the chemists write in Science, the de novo-modeled catalysts proved effective without the use of directed evolution.
Cyclopropanation and insertion reactions into element-hydrogen bonds can be carried out using organometallic catalysts. Typically, in such a catalyst, the metal is bound to a chiral ligand, which ensures that the reaction product is formed as a single optical isomer. With this approach, selecting a suitable ligand can be difficult, and chemists must explore many different catalysts in search of the most effective one.
Another approach to asymmetric catalysis involves coupling organometallic catalysts to proteins. In this case, instead of chiral ligands, the protein structure is responsible for selectivity. If the active site structure is chosen correctly, the amino acid residues will "direct" the reaction in the active site so that only one optical isomer of the product is formed. However, if such a protein catalyst is not performing well, its structure can be optimized using directed evolution. We discussed how this works in the article "Playing God."
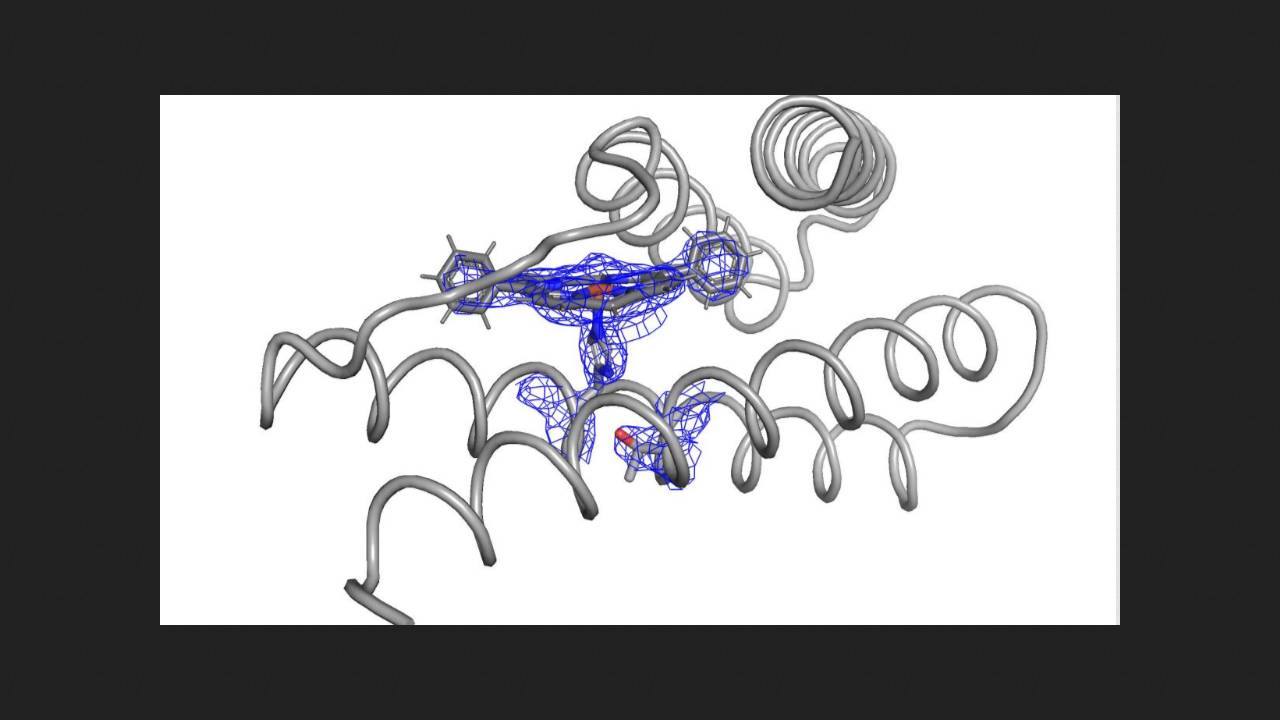
It was this second approach that was recently employed by chemists led by William F. DeGrado of the University of California, San Francisco. Their idea was to model protein catalysts for the reactions of cyclopropanation of alkenes and insertion into silicon-hydrogen bonds.
First, the chemists took a known catalytically active protein with a diphenylporphyrin cofactor (which forms a complex with an iron ion) and tested it in the cyclopropanation of styrene with ethyl diazoacetate. The product was obtained with a yield and an enantiomeric excess of 40 percent. The scientists then hypothesized that the protein's structure prevented reagents from easily penetrating the active site, leading to the low yield.
To model more effective catalysts based on this protein, the scientists used several machine learning algorithms, including one previously developed in their lab. As a result, they obtained a set of several dozen protein structures, from which they selected ten with the most well-defined active site structures and expressed them in bacteria. Most of the resulting proteins proved to be effective catalysts for styrene cyclopropanation. The best result achieved was a quantitative product yield and enantiomeric excess of approximately 99 percent.
The chemists then used the same method to model protein structures to catalyze the insertion of diazo compounds into silicon-hydrogen bonds. This time, however, instead of diphenylporphyrin cofactor, they chose protoporphyrin IX, which acts as a heme precursor in cells. The chemists again expressed the selected proteins in bacteria, and they proved to be effective catalysts. Furthermore, the scientists were able to further enhance the enantioselectivity of the insertion by directed evolution of the resulting catalysts in living cells. This was possible because the scientists chose the cellular metabolite protoporphyrin IX as a cofactor.
Thus, the chemists succeeded in obtaining a set of protein catalysts for two reactions. However, despite the high efficiency of the resulting catalysts, minor changes in the structure of the starting materials often resulted in a decrease in yield and enantioselectivity.
Previously, we reported on how chemists used yeast lipase in combination with a ruthenium catalyst for the enantioselective synthesis of macrocycles.