Russian scientists have discovered a previously unknown bacteriophage, Sxt1, in a therapeutic phage cocktail for treating bacterial infections. It is capable of infecting a number of wild strains of E. coli resistant to phages from the same family. After comparing Sxt1 with related phages T3 and T7, the scientists discovered that the secret to its effectiveness lies in the structure of its tail fibers. The results of the study were published in the journal Viruses.
One solution to the problem of bacterial resistance to antibiotics is to treat bacterial infections with bacteriophages—viruses that infect bacteria. However, phages are highly specific and can only infect certain strains of a single bacterial species. Therefore, phage cocktails—mixtures of different phages active against different strains of one or more bacterial species—are used in therapy. However, due to the lack of data on the clinical efficacy of therapeutic phage cocktails, their use is limited.
A team of scientists from the Skoltech Metagenome Analysis Laboratory, led by Artem Isaev, studied the composition of the therapeutic phage cocktail "Sextaphage (polyvalent pyobacteriophage)," produced by the Russian company Mikrogen, and isolated a new bacteriophage, Sxt1, which was able to infect a wide range of E. coli strains, including some that are resistant to other, related phages.
The polyphage cocktail "Sextaphage" contains phages isolated from Staphylococcus spp., Streptococcus spp., Proteus (P. vulgaris, P. mirabilis), P. aeruginosa, Klebsiella pneumoniae, and enteropathogenic E. coli. In the study, the scientists used the E. coli strain BW25113 as a host. The Sxt1 phage from the Autographiviridae family was isolated from the cocktail. It turned out to be a close relative of the E. coli phages T3 and T7. Phages from this family possess RNA polymerase, which transcribes a number of phage genes. The immediate-early genes (genes transcribed immediately after infection of a bacterial cell) of Sxt1, early genes (involved in DNA metabolism and phage genome replication), and some late genes (encoding structural components of phage virions, their packaging, and bacterial lysis) were almost completely identical to the genes of phage T3. Differences between Sxt1 and related phages were found in the genes encoding tail fibers and two internal proteins and are, according to scientists, the result of phage recombination, presumably with the Berlinvirus phage.
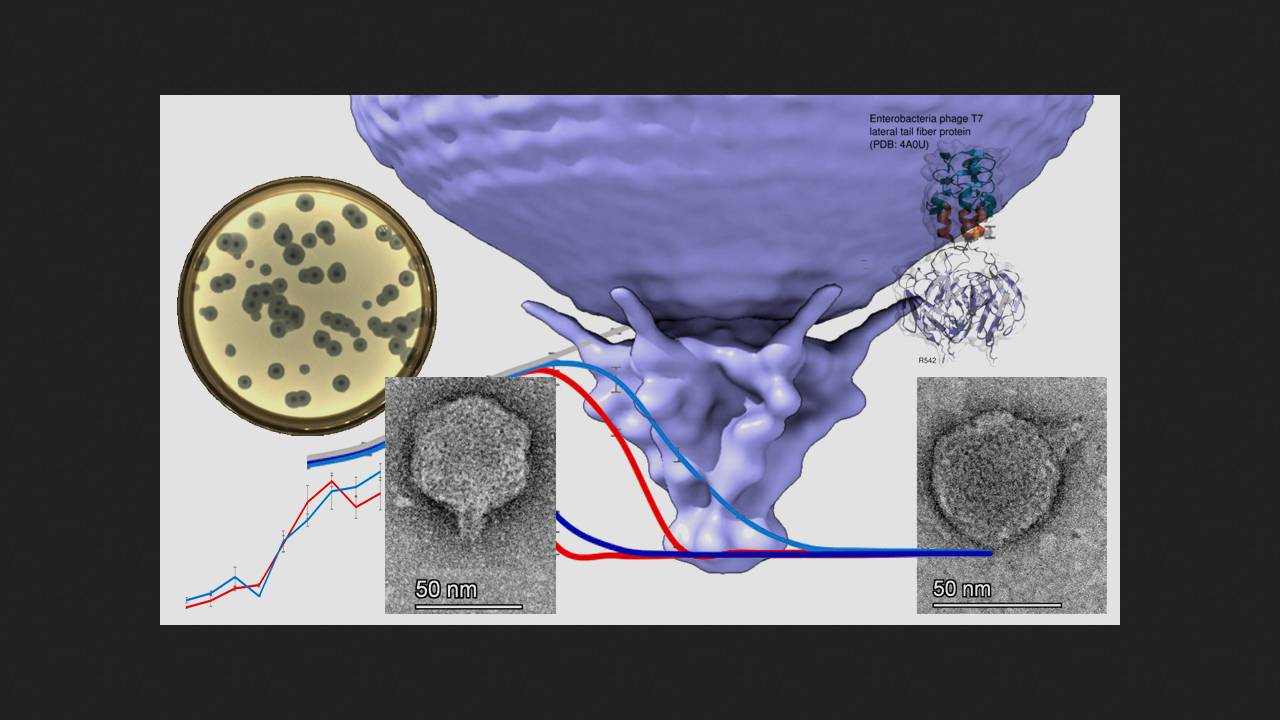
Исследователи построили филогенетическое древо бактериофагов подсемейства Studiervirinae, куда входит Sxt1, определили его предполагаемое происхождение и пришли к выводу, что секрет его исключительности кроется именно в белке латеральных хвостовых фибрилл. Фаги из семейства Autographiviridae для прикрепления к бактериальной клетке используют шесть хвостовых фибрилл, каждая из которых состоит из тримера этого белка. На концах тримера имеются участки, контактирующие с поверхностью клетки. N-концевой домен необходим для прикрепления к капсиду, длинный пирамидальный домен включает в себя бета-листы от каждой из трех молекул, входящих в состав тримера, за ним расположен короткий альфа-спиральный линкер и концевой домен, отвечающий за взаимодействие с бактериальной клеткой. Пирамидальный домен Sxt1 содержал шесть вставок длиной более пяти аминокислот, причем некоторые из вставок образовывали дополнительные бета-слои. Линкер также включал в себя дополнительную вставку из 27 аминокислот. В итоге за счет вставок хвостовая фибрилла Sxt1 длиннее, чем у Т3 и Т7. С помощью программы AlphaFold2 ученым удалось смоделировать концевой домен Sxt1 с альфа-спиральным линкером и часть пирамидального, и оказалось, что она только на 43 процента идентична аналогичной области фага Т3 и на 54 процента — фага Т7. При этом хвостовые фибриллы Sxt1 распознают иной набор рецепторов бактериальной клетки по сравнению с Т3 и Т7.
Для оценки специфичности нового фага исследователи высеяли фаги Sxt1, T7 и T3 на распространенные штаммы E. coli: BW25113, MG1655, BL-21, B, C, DH5α, HS и Nissle1917, а также на штаммы F+ BW39773 и KD263, которые способны ингибировать T7 из-за наличия системы абортивной инфекции PifA. Устойчивыми ко всем фагам оказались только штаммы HS и Nissle1917. Все остальные штаммы E. coli фаг Sxt1 успешно инфицировал. При этом Т3 не смог заразить штаммы линии К12, а Т7 потерпел неудачу со штаммами с системой PifA.
To determine the host range of the Sxt1 phage, scientists tested its interactions with different strains of E. coli. E. coli bacteria are divided into serogroups based on the structure of their O-antigen—a highly variable polysaccharide on the outer membrane of their cell wall that allows bacteria to evade the protective effects of the adaptive immune system. The researchers hypothesize that O-antigens are one of the main obstacles to phage recognition. Therefore, the higher the specificity of a phage to bacterial O-antigens, the more effective it is for phage therapy. The researchers used the ECOR collection—a set of natural E. coli isolates with divergent O-antigen types—to test the effects of phages Sxt1, T3, and T7 on different strains. The Sxt1 phage was able to infect 15 of the 72 strains in the collection (20 percent), including all bacteria susceptible to T7 and/or T3, and an additional 7 strains resistant to these phages. The researchers then analyzed the phage's effect on the ECOR50 strain, which is sensitive to Sxt1 but not to T7 or T3, and found that only Sxt1 was able to bind to its surface. This suggests that Sxt1's effectiveness stems from its ability to recognize bacterial receptors, not from increased resistance to the cell's defenses.
Scientists have concluded that the Sxt1 phage, with its extended host range, is more effective than its relatives, the T3 and T7 phages. It has great potential for therapeutic use against E. coli infections, and assessing its specificity could become a standard procedure for characterizing bacteriophages.
In recent years, in the fight against antibiotic resistance, there has been an active search for new bacteriophages and the development of methods for modifying them. In 2021, American scientists integrated the CRISPR/Cas9 system into a bacteriophage, allowing it to target a specific strain of E. coli. That same year, Portuguese scientists created synthetic bacteriophages that target Pseudomonas aeruginosa.